- Home
- About ACA
- Publications & Resources
- Programs
- Annual Meeting
- Membership
- -History-
- ACA Video Library
ACA Early Career Scientist Spotlight-24
Personal StatementI am currently working in the RNA Structural Biology field, focusing on the structures and functions of various viral RNAs and related proteins. I'm particularly interested in understanding the interactions between viral RNAs and both viral and host proteins. During my master’s program at MS State, I developed a strong interest in Biochemistry, thanks in part to the encouragement of some friends. It was during doing my rotation with Dr. Koirala that I was inspired to join his lab. He shared several fascinating stories about crystallizing viral RNA domains and solving structures. I'm proud to say that the first crystal structure I solved was the Coxsackievirus B3 (CVB3) RNA replication platform.
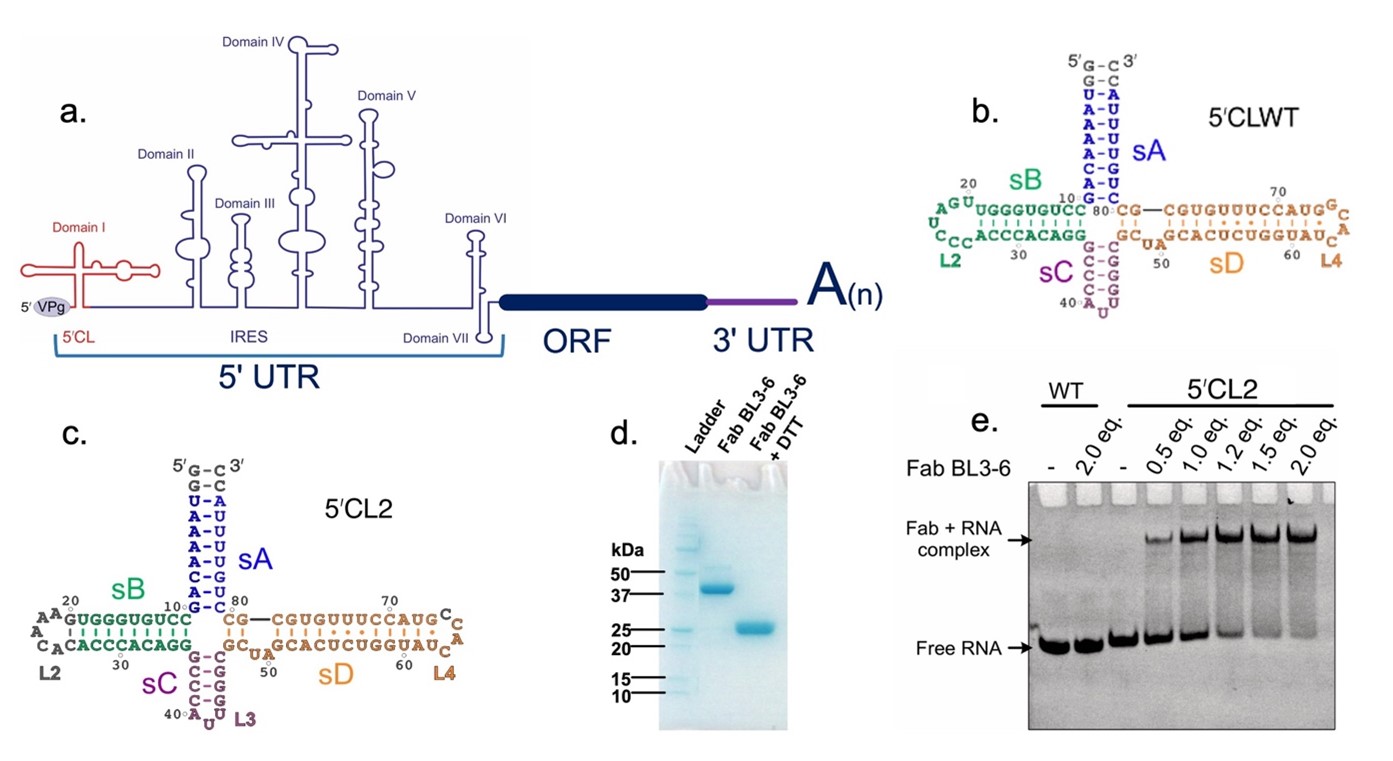
CVB3 represents a major human pathogen that causes acute and chronic viral pancreatitis and myocarditis. It belongs to the enterovirus genus of the Picornaviridae family. Every year, there are about ten to fifteen million enterovirus infections and more than ten thousand hospitalizations take place in the USA. These pathogenic viruses are responsible for human diseases such as the common cold, poliomyelitis, acute flaccid paralysis, and myocarditis. Nevertheless, except for the poliovirus (PV), no vaccine or effective treatment is available. These viruses get mutated and frequently change their forms. They may emerge again in a different form. The CVB3 (+)-sense RNA genome can be divided into three main parts: the 5' untranslated region (5' UTR), the open reading frame (ORF), and the 3' untranslated region (3' UTR). The 5' UTR (Fig. 1a) contains two types of modular domains, with domains II-VII known as internal ribosome entry sites (IRES). At the extreme 5' untranslated region of the genome, there is a cloverleaf-like (CL) RNA domain that forms an essential platform for viral genome replication (Fig.1a). This domain interacts with the host PCBP2 and viral 3C proteins through its stem-loop subdomains, B (sB) and D (sD), respectively. Although the 5'CLs are crucial and highly conserved among enteroviruses, the high-resolution structures of these domains remain unknown.
As our general workflow, we focus on RNA viruses and proceed with designing RNA constructs and growing crystals, which we then send to a synchrotron facility for diffraction data collection. Once we obtain high-quality diffraction data, we use phasing, modeling, and refinement techniques to solve the 3D structure of the RNA. In our lab, we use Fab-assisted RNA crystallography, where Fab is used as a chaperone, which helps crystallization and can be used as a molecular replacement model to get phase information. Specifically, Fab BL3-6, in this case, binds to an RNA epitope 5' GAAACAC 3'. Thus, we grafted the RNA epitope into the wild-type RNA construct (Fig. 1b) to obtain a crystallization construct (replaced L2, colored gray in Fig.1c). We synthesized and purified the RNA and the Fab BL3-6 (Fig. 1d). Next, we performed crystallization trials after a good RNA-Fab BL3-6 binding (Fig. 1e). Beautiful crystals grew from the trial and diffracted at 1.9 Å. We solved the first-ever intact crystal structure of an enteroviral RNA replication platform. The crystal-derived RNA structure looks like an H rather than widely known Cloverleaf-like shape (Fig. 2a). In the Fab-RNA complex, Fab BL3-6 is bound in the sB as designed. The sA stacks on sD, and sB stacks on sC, and there is a four-way junction with no nonpaired nucleotides within the junction connecting strands. The structure revealed an unprecedented Py-Py interaction between the sC and sD (Fig 2a, 2b). Then, we also confirmed that this tertiary interaction also occurs in solution with NOE NMR studies (Fig. 2c).
Now, we asked the question, why are the Py-Py tertiary interactions important? To find out we designed 5'CL2-A40U mutation constructs, purified CVB3 3C protein (Fig. 2d) and tested the binding with 3C protein in ITC, and found that both crystallization constructs, 5'CL2 and 5'CL2-A40U mutation RNA construct, were binding binds with similar affinity (Fig. 2e). Thus, we concluded that this interaction is not essential for 3C protein binding. However, it may be necessary for poly-C binding protein (PCBP) binding. Then, I purified the human PCBP2 protein (Fig. 2f) and tested its binding with 5'CLWT, 5'CLWTSP (SP-spacer), and 5'CLSPA40U constructs using electrophoretic mobility shift assay (EMSA). The result suggests that it may be essential for PCBP binding (Fig. 2g). Further experiments are required to understand the effect of tertiary interaction better. Overall, the secondary structure is H-type instead of the previously predicted cloverleaf, and unprecedented Py-Py interaction between sC and sD is important for PCBP2 binding. With this being the first high-resolution resolution structure of the enteroviral 5'CLs, our research will pave the way for future studies into the mechanism of enteroviral genome replication and developing antiviral drugs that target this RNA-centric platform. Our lab focuses on the structural elucidation of enteroviral replication and IRES-based translation. Hasan Al Banna and Manju Ojha are the other two graduate students in the lab, and our group also includes several undergraduate and high school students. I used Microsoft PowerPoint to create my poster, which was then reviewed by my advisor, Dr. Koirala. While there are other software options available, such as Adobe Illustrator and CorelDRAW, I found PowerPoint to be the easiest and most accessible for students. I am passionate about various activities, including gardening, biking, hiking, reading, cooking, playing outdoor games, and traveling. Additionally, I'm actively involved with a non-profit organization, where I find great joy in assisting the cook and distributing free meals to homeless individuals in downtown Baltimore. |